CPIC Guideline Update for TPMT, NUDT15 and Thiopurines
The Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Thiopurine Dosing Based on TPMT and NUDT15 Genotypes: 2025 Update has been published in Clinical Pharmacology and Therapeutics (PMID:41618934). The guideline contains multiple updates to the previous recommendations for adjusting the starting doses of azathioprine, mercaptopurine and thioguanine according to the combination of TPMT and NUDT15 phenotypes derived from genotypes and the specific indication (i.e. malignant or nonmalignant condition). The guideline update emphasizes a greater reduction of dose for compound TPMT/NUDT15 intermediate metabolizers.
The guideline manuscript, supplement, all supporting tables including allele definition, allele function, gene CDS, pre- and post-test alerts, etc., and links to ClinPGx guideline annotations with the accompanying genotype/phenotype selection tool to retrieve specific recommendations, can be found on the ClinPGx website on the guideline page. Updated information is also available in the CPIC DB.
As part of the guideline process, CPIC's TPMT and NUDT15 PCEP groups re-evaluated and updated the function assignments for the respective star alleles, with each gene having at least one allele assigned decreased function. Previously all TPMT and NUDT15 alleles were normal function, no function or unknown/uncertain function. The addition of decreased function alleles required updated mapping from allele function to phenotype for both TPMT and NUDT15.
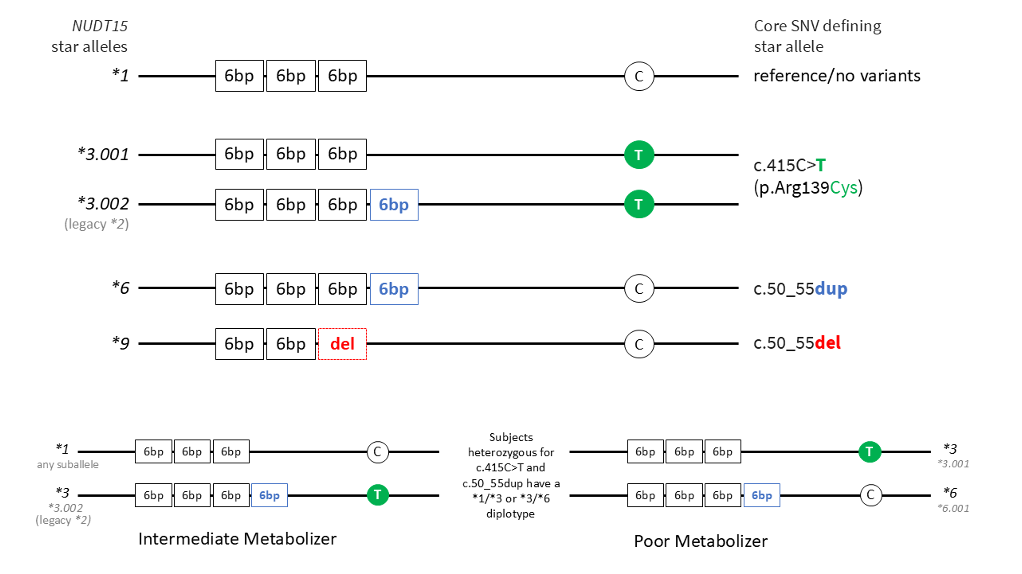
During the function evaluation process, PharmVar's NUDT15 expert panel determined that NUDT15*2 and *3 should be consolidated under the same star number as both have c.415C>T (p.R139C) which eliminates enzymatic activity. The NUDT15*2 allele has been removed from ClinPGx and CPIC materials and DB reflect the transition of *2 to a *3 suballele.
The c.415C>T variant can occur without (*3.001) and with (*3.002) the 6 base pair GAGTCG insertion (also referred to as c.50_55dup), and the insertion can also occur without c.415C>T (*6). The figure below provides a graphical overview of these NUDT15 star alleles. Because the single nucleotide variant and the insertion variant can be found in different combinations on the same gene copy or different gene copies, the phase of the variants (i.e., whether the variants are in cis or in trans) is important. As shown in the bottom panel of the figure, for a patient with a heterozygous test result for both c.415C>T and c.50_55dup without any information regarding variant phase, the genotype is ambiguous. The patient has either a NUDT15*1/*3 (intermediate metabolizer) or *3/*6 (poor metabolizer) diplotype with the former having a *3.002 suballele and the latter having a *3.001 suballele. As the predicted phenotypes for NUDT15*1/*3 and *3/*6 differ, it is important to be aware of this ambiguity. Currently, there is no standardized approach to reporting ambiguous diplotypes when phase is unknown. Additionally, if a pharmacogenetic test does not interrogate the GAGTCG repeat, NUDT15*6 and *9 alleles are not detected and are ‘defaulted’ to *1, while *3.002 will be identified as *3 due to having c.415C>T. Please refer to the PharmVar NUDT15 Read Me document for more information.